Using DrugBank Conditions Data
Introduction
In DrugBank, conditions cover everything that can happen in the human body, such as the diseases a drug can treat, the adverse effects patients may have in response to a given drug, and even the characteristics of patients for whom drug indications or warnings are applicable. Because of how they are used, our conditions ontology will appear in many different datasets. Therefore, it is important to understand their relevance and how they can be used to leverage DrugBank’s data.
How are conditions data organized?
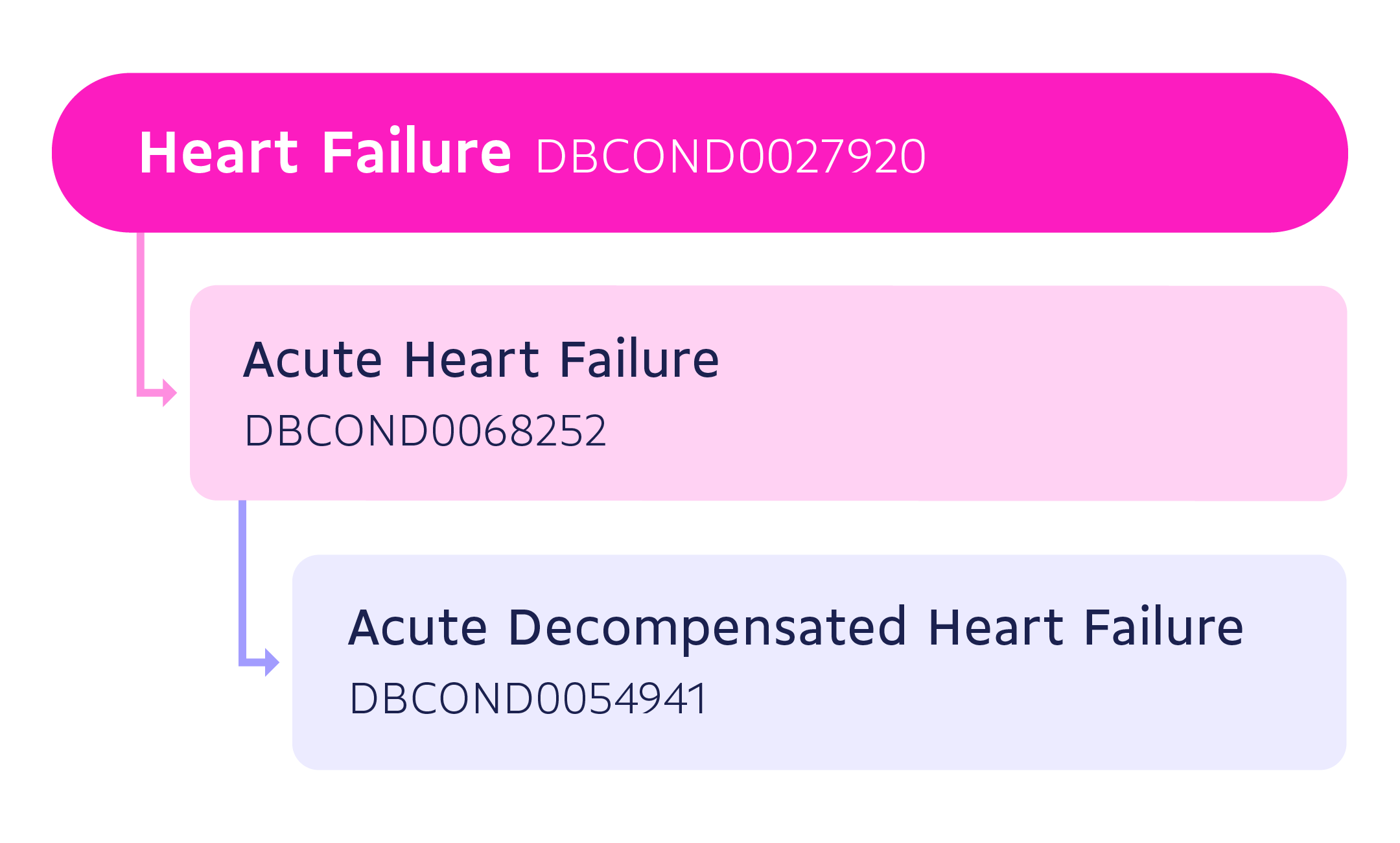
Conditions in our database can be identified by their DrugBank condition ID (an alphanumeric code with the prefix DBCOND). Data follows a hierarchical, or tree-like, structure based on parent-child relationships, where an element higher in the hierarchy (parent condition) is inclusive of all conditions lower in the hierarchy (child conditions). Parent and child conditions are closely related to each other, and a child is a more specific or detailed form of a parent. For example, Acute Heart Failure (AHF) (DBCOND0068252) is a child of Heart Failure (DBCOND0027920), and Acute Decompensated Heart Failure (ADHF) (DBCOND0054941) is a child of Acute Heart Failure (AHF).
The connectivity and usability of conditions data
The value of DrugBank data resides in its connectivity. Within DrugBank, conditions data is connected to various datasets, including structured indications, contraindications, boxed warnings, adverse events, and allergy data. This connectivity extends to some of the most commonly used external ontologies, including ICD-10, SNOMED, and MedDRA, which improves the usability of our conditions data. Moreover, due to the variability between different terms and the risk of data duplication, conditions data in DrugBank relies on the use of synonyms and preferred terms.
Synonyms avoid repetition between conditions and enable better data matches. By synonymizing terms such as Retinal Dystrophy (DBCOND0009607) and Dystrophy, Retinal (DBCOND0135695) or Dry Mouth (DBCOND0038951) and Xerostomia (DBCOND0023846), it is possible to query data using equivalent terms. However, for ease and consistency, a single term (the preferred term) will be shown across all datasets. To make these associations, a relational table between preferred terms and the different synonyms of each condition is included in our downloads data.
How can this be used?
The use of external mapping connectivity and hierarchies expands the capabilities of DrugBank’s conditions data. Conditions terminology may change from one source to another, and in some cases, a single term will not be able to encompass the data required for a specific search. Therefore, the use of external mapping and hierarchies will improve flexibility regardless of the dataset being queried.
Why does this matter?
The overall objective of taking a drug from the drug discovery and development stage to the point where it is used in clinical settings is to benefit a segment of the population with similar characteristics. The patient is at the center of this process, and each condition they present with or develop is highly relevant. Therefore, understanding the role that conditions play in each situation, and getting an insight into how they are connected with other types of data will help users of DrugBank to solve many questions revolving around patients and diseases. For instance, the conditions data in DrugBank can be used for disease-centric approaches in drug discovery, or to better understand the different mechanisms involved in a condition.
External Mappings
Use of external mappings
Conditions data in DrugBank is able to connect to external ontologies including ICD-10, SNOMED, and MedDRA, which each possess their own hierarchy. These external ontologies are widely used for different purposes in clinical and industry settings; therefore, having the ability to match DrugBank conditions to the terms in these ontologies increases usability.
Query example
The query below shows an example where all indications matching the ICD-10 code that corresponds to Osteoporosis in females and males (c/M81.0) are pulled:
SELECT DISTINCT d.drugbank_id AS 'DBID', d.name AS 'Drug', si.kind AS 'Indication Kind', c.drugbank_id AS 'DBCOND', c.icd10_id AS 'ICD-10', c.title AS 'Indication Disease' FROM structured_indications si JOIN drugs d ON si.drug_id=d.id LEFT JOIN indication_conditions ic ON ( si.id=ic.indication_id AND ic.relationship='for_condition' ) LEFT JOIN conditions c ON ic.condition_id=c.id WHERE c.icd10_id = 'c/M81.0';Copy to clipboard
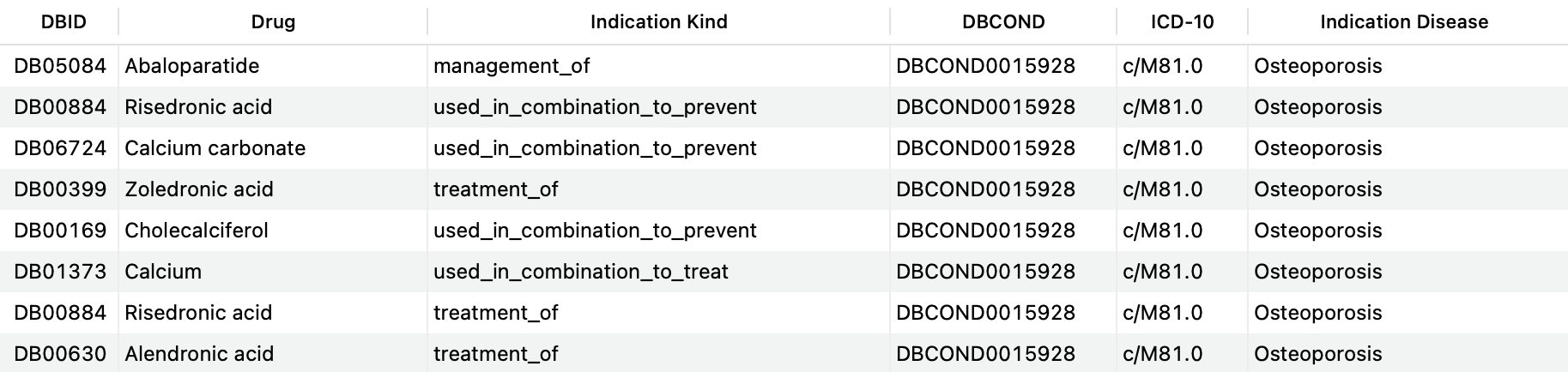
As a result, we obtain a table that includes the drugs used to treat, prevent or manage this condition.
Keep in mind that since we are using SQL, we can take advantage of wild cards to expand our search. For example, looking for 'c/J45%' will return detailed information on the different conditions related to Asthma, under the ICD-10 J45 category.
How are the tables joined?
For this query, we need to join the drugs table to the structured_indications table using the drug ID, a common identifier used to gather information about each drug and its indications. The structured_indications table is then joined to the indication_conditions table through the indication ID to see which condition is associated with each indication. Afterwards, the indication_conditions table is joined to the conditions table to get the details about each condition, including their identifiers or DBCOND as well as their external identifiers.
Conditions Hierarchy
Use of hierarchies
The hierarchies in our conditions data enable DrugBank users to match conditions using different levels of specificity, which expands their search and increases the probability of finding the results they need.
Query example
For instance, a user may be interested in getting a list of all drugs used to treat, prevent or manage Osteoporosis (DBCOND0015928) as well as subtypes (direct child concepts) of this condition, such as Postmenopausal Osteoporosis (DBCOND0030339). The query below is able to obtain this information:
SELECT DISTINCT d.drugbank_id AS 'DBID', d.name AS 'Drug', si.kind AS 'Indication Kind', c_ch.drugbank_id AS 'DBCOND', c_ch.title AS 'Indication Disease' FROM structured_indications si JOIN drugs d ON si.drug_id=d.id LEFT JOIN indication_conditions ic ON ( si.id=ic.indication_id AND ic.relationship='for_condition' ) LEFT JOIN conditions c_ch ON ic.condition_id=c.id LEFT JOIN condition_children cc ON c_ch.id=cc.child_preferred_term_id LEFT JOIN conditions c_p ON cc.parent_preferred_term_id=c_p.id WHERE c_ch.drugbank_id = 'DBCOND0015928' OR c_p.drugbank_id = 'DBCOND0015928';Copy to clipboard
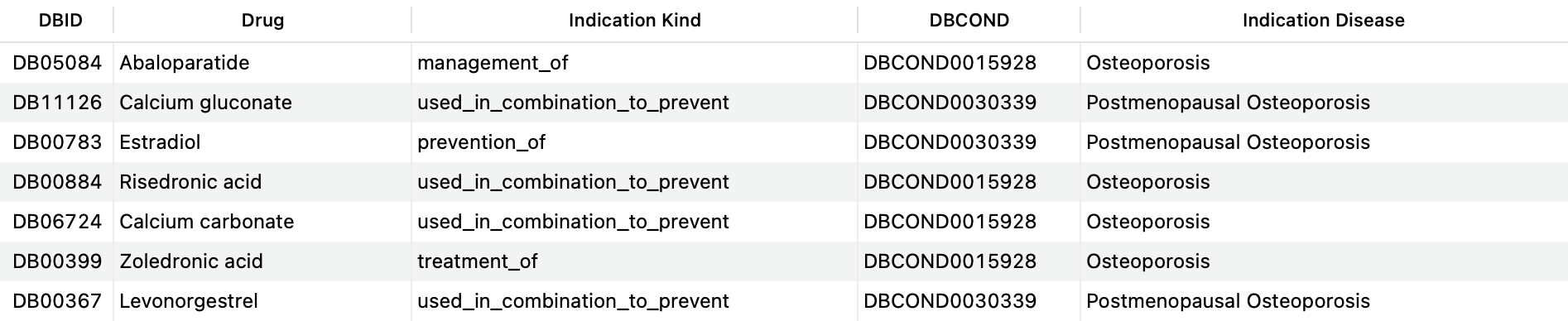
This query is asking the system to return all indications where osteoporosis or a child of osteoporosis is the disease a drug is indicated for.
How are the tables joined?
This query also relies on data that can be found in the indications dataset, and uses the structured_indications and indication_conditions tables. Similar to the query above, we also need to join the drugs and conditions table to gather information about drugs and conditions. However, in this case, we also need to use the condition_children table (which includes hierarchy information) and associate it with the conditions table once again.
Conclusion
Conditions help us understand phenomena directly related to patients, such as the positive and negative effects of treatments, as well as their circumstances and pre-existing diseases. Dealing with conditions data can be challenging due to the variability in terminology across different sources and ontologies, which can introduce data conflicts and inconsistencies. In order to avoid this, DrugBank’s conditions data follows a hierarchical structure and is associated with some of the most commonly used external ontologies. Moreover, since it appears in many different datasets, the conditions data in DrugBank can be used to draw connections required to solve some of the questions in drug discovery and clinical research.