Using DrugBank Target Data
Introduction
DrugBank contains a wealth of information related to direct interactions between drugs and biomolecules, namely proteins. Within DrugBank, these interactions are referred to as “bonds.” There are four types of bonds, denoted by the 'type' column of the bonds table:
| Bond Type | Description |
|---|---|
| TargetBond | The drug binds to the biomolecule and affects its function; often, this is directly related to the drug's mechanism of action. |
| EnzymeBond | The drug binds to an enzyme or otherwise affects its function, including by inducing/inhibiting the enzyme. |
| CarrierBond | The drug binds to a carrier protein in the plasma. |
| TransporterBond | The drug binds to a transmembrane protein transporter or otherwise affects its function, including induction/inhibition of transport. |
Why do these matter?
The ability to develop drugs and understand their clinical utility for individual patients is increasingly reliant on having sophisticated knowledge of how drugs interact with specific biomolecules. The binding of drugs to proteins and other biomolecules can elicit alterations in their function, which forms the basis for therapeutic efficacy (pharmacodynamic effects). Similarly, drug binding to enzymes, carriers, and transporters form the basis for an understanding of a drug's pharmacokinetic properties, or the way in which a drug is distributed throughout the body. Understanding a drug's pharmacodynamics and pharmacokinetics is crucial in the modern chemotherapeutic landscape.
How does DrugBank make it easier to leverage this information?
Molecular interactions between drugs and biomolecules are determined through a variety of techniques, both biochemical and computational. As a result, the largest resource for such data remains the body of biomedical research. Extracting these valuable insights from such a large volume of data is challenging; manual assessment by individual researchers is laborious and time-consuming, while methods relying solely on algorithmic or machine learning (ML)-based approaches can be error-prone.
DrugBank combines automated approaches based on natural language processing with expert evaluation by an in-house team of biomedical experts. As such, our bonds dataset is both comprehensive and accurate, with extensive references to published literature to allow for independent validation. This dataset is also highly structured, which makes it easy to query and use as input to algorithms or ML models.
What can be done using bonds
Our structured bonds dataset makes it easy to investigate the molecular interactions of a drug or group of drugs. More complex questions can also be answered due to the extensive connectivity of our data: interaction with specific biomolecules can be tied to efficacy, adverse reactions, and many other phenomena. Though we provide several examples below, many more questions can be answered through appropriate querying.
Working with Drugs
The simplest way to leverage this information is to retrieve bonds of a given type related to a drug of interest:
SELECT d.drugbank_id, d.name, b.biodb_id, b.pharmacological_action FROM bonds b JOIN drugs d ON b.drug_id=d.id WHERE d.name = 'Acetaminophen' AND b.type = 'TargetBond';Copy to clipboard
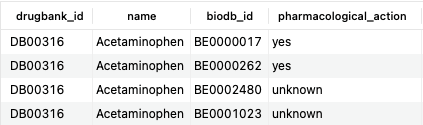
The above query returns all target bonds associated with acetaminophen, a common analgesic. The first two interactions are associated with acetaminophen's mechanism of action ('pharmacological_action' is yes), while the latter two may or may not be ('pharmacological_action' is unknown).
How can this be used? (scenario and explanation)
This kind of query is useful to know the targets for an individual drug. It might be the case that additional biochemical studies are desired around an individual drug-biomolecule interaction, for example, to understand the exact mechanism of action better. Alternatively, a novel molecule may have been identified that is similar to an existing drug; in this case, the targets represent reasonable starting points to investigate the binding affinities of the new molecule.
How are we joining the tables?
In this call, we are joining the drugs table and the bonds table through a common identifier called the drug id. This column is present in the drugs table as 'id' as it belongs to the drugs table. In the bonds table, the relation id is called the 'drug_id'; this is because the bonds table is a separate entity from the drugs table.
Working with Drugs and Bioentities
While the above query is useful, it would be nice to know which targets these are. Note the 'biodb_id' column, whose entries contain seven-digit identifiers prefixed by “BE.” These are “bioentity IDs.” In DrugBank, bioentities are standard entries to represent biological molecules. Protein bioentities (the most common kind of bioentities) are associated with a UniProt identifier, providing a link to an external resource with much more information. By joining the bio_entities and bio_entity_components tables, we can get much richer information:
SELECT d.drugbank_id, d.name, b.biodb_id, be.name, bec.component_id, b.pharmacological_action FROM bonds b JOIN drugs d ON b.drug_id=d.id JOIN bio_entities be USING (biodb_id) JOIN bio_entity_components bec USING (biodb_id) WHERE d.name = 'Acetaminophen' AND b.type = 'TargetBond' AND be.kind = 'protein';Copy to clipboard

Now we can identify the two targets related to the mechanism of action as prostaglandin G/H synthase 1 and 2; prostaglandins are known to regulate pain and inflammation. Using the UniProt IDs present in the 'component_id' column, it is possible to learn much more about these synthases.
How can this be used? (scenario and explanation)
This query is useful in the same ways as the query above; the main advantage here is the human readability factor. It is possible, for example, to individually query the bio_entities table with each bioentity ID. However, this is inefficient and requires jumping between tables. In addition to identifying each target by name, this query also includes UniProt identifiers, which can be used to learn more about each target.
How are we joining the tables?
In this query, we are joining four tables together. First, we will join the drugs and bonds tables, which is what we saw in the first query of the article. This is done through the drug ID found as 'id' in the drugs table and 'drug_id' in the bonds table.
From there, we join the bio_entities table that translates the bio entity id ('biodb_id', example: BE0000017) found in the bonds table to a human-readable name. To learn more about what a bio entity is, click here. In both the bonds table and the bio_entities table, the identifier will be in the column titled 'biodb_id.'
Last but not least, we are also joining the bio_entity_components and bio_entities tables via the 'biodb_id.' We do this to be able to pull the component id, which will represent a UniProt ID. This ID is crucial to bond information as it can be used for referencing in UniProt and also allows you to join to other tables like polypeptides. This can provide you with additional insight, such as the name of the gene that gives rise to the protein the drug acts on.
Working with Categories
Often, you may be interested in a class of drugs rather than any single drug. By combining DrugBank categories with target information, it is possible to identify common targets across a range of medications:
SELECT b.biodb_id, be.name, bec.component_id, COUNT(DISTINCT d.name) AS target_count FROM bonds b JOIN bio_entities be USING (biodb_id) JOIN bio_entity_components bec USING (biodb_id) JOIN drugs d ON b.drug_id=d.id JOIN drug_categorizations dc ON d.id=dc.drug_id JOIN categories c ON dc.category_id=c.id WHERE c.title = 'Penicillins' AND b.type = 'TargetBond' AND b.pharmacological_action='yes' GROUP BY be.name ORDER BY target_count DESC;Copy to clipboard
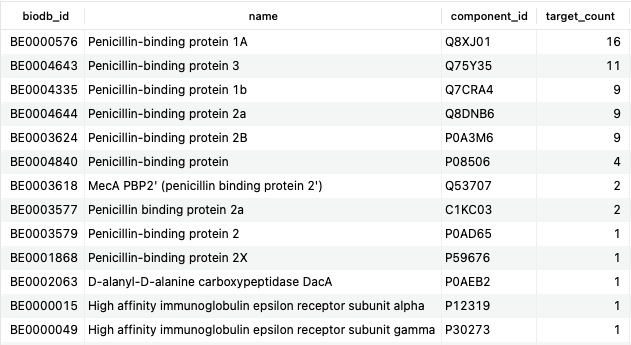
Here, we have used several additional SQL clauses (COUNT, GROUP BY, and ORDER BY) to obtain a count of targets related to the mechanism of action of penicillins. Unsurprisingly, the majority of these are penicillin-binding proteins, but the preponderance of 1A and 3 are interesting to note.
How can this be used? (a scenario and explanation)
This query goes beyond querying individual drugs to entire categories of drugs; a similar approach could be used for other sets of drugs that don't exactly correspond to one of our categories as well. By comparing targets across a group of drugs, it is possible to understand more about their functionality, both in terms of similarities and differences. In a hypothetical example, we could interrogate a group of drugs that all achieve a similar therapeutic effect. If the list of shared targets is small, it suggests a limited number of options for therapeutic intervention. However, if the list is large, it may suggest some promiscuity; if the effect is modulated by a large pathway, for example, it may be possible to target one of several pathway members to achieve a similar downstream effect.
How are we joining these tables?
We are again joining tables related to bio entities (see Working with Drugs and Bioentities). In order to filter the set of drugs by categories, we first join the drug_categorizations table by looking for rows where the 'drug_id' is the same as the 'id' in the drugs table. The drug_categorizations table holds information into which category(ies) each drug is classified. From here, we can now join the categories table itself by equating the 'category_id' to the 'id' of each row in the categories table.
Focussing on Individual Biomolecules
Up until now, we have been focused on going from a drug or category to relevant targets. It is also possible, and often of interest, to go the other way, i.e., from a specific biomolecule to all relevant drugs. As an example, let's consider the case of liver enzymes involved in drug metabolism and attempt to identify drugs that induce the important enzyme CYP3A4:
SELECT d.drugbank_id, d.name, be.name, b.inducer FROM bonds b JOIN bio_entities be USING (biodb_id) JOIN bio_entity_components bec USING (biodb_id) JOIN polypeptides p ON bec.component_id=p.uniprot_id JOIN drugs d ON b.drug_id=d.id WHERE p.gene_name = 'CYP3A4' AND b.inducer = '1';Copy to clipboard
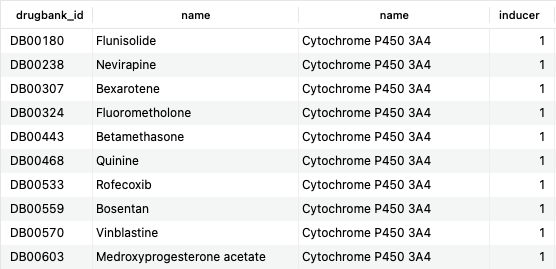
This query returns 209 rows, the first 10 of which are shown here. The result of this query corresponds to the DrugBank category “Cytochrome P-450 CYP3A4 Inducers.” Still, by using SQL, it is possible to interrogate further the data (what if you were interested in drugs that were either an inducer or an inhibitor?).
How can this be used? (a scenario and explanation)
By inverting the query, i.e., focusing on the biomolecule(s) rather than the drug(s), we can understand the types of molecular scaffolds capable of interacting with an individual biomolecule. Extracting the chemical structures for further analysis may reveal crucial substructures, functional groups, or other entities that affect binding or efficacy. Further filtering the results of these interactions (e.g., for inducers, inhibitors, etc.) can allow comparisons of molecules that all bind but whose binding has different effects. Such data can help to inform rational drug design.
How are we joining these tables?
This query is again similar to those already discussed. The change here is the addition of the polypeptides table, which is joined through its 'uniprot_id' column to the 'component_id' of the bio_entity_components table. This gives access to gene names, which are usually shorter and follow a more standardized naming convention, compared to full-length protein names.
Tying in Other Datasets
While DrugBank bonds data is useful on its own, there is great value in connecting it to other areas of our data. For example, we can interrogate the adverse effects of drugs associated with one or more targets:
SELECT co.drugbank_id, co.title, COUNT(DISTINCT d.id) AS AE_count FROM structured_adverse_effects sae JOIN adverse_effect_conditions aec ON sae.id=aec.adverse_effect_id JOIN conditions co ON aec.condition_id=co.id JOIN drugs d ON sae.drug_id=d.id JOIN bonds b ON d.id=b.drug_id JOIN bio_entities be USING (biodb_id) WHERE be.name LIKE '%5-hydroxytryptamine receptor%' AND aec.relationship = 'effect' AND b.type = 'TargetBond' GROUP BY co.drugbank_id ORDER BY AE_count DESC;Copy to clipboard
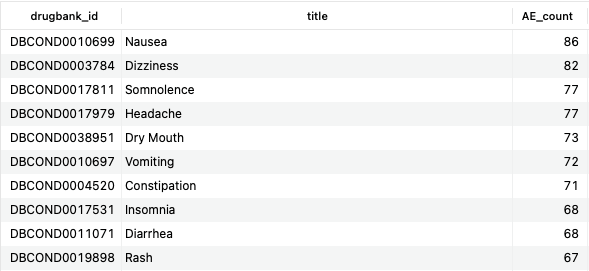
Here, we show the common side effects among drugs that target members of the 5-hydroxytryptamine (5-HT) receptor family, otherwise known as serotonin receptors. The majority of the top hits are consistent with the modulation of serotonergic pathways, as one would expect.
How can this be used? (a scenario and explanation)
Often, we may be interested in a target or group of targets in terms of their clinical implications. This could include their intended use (indication), interactions with food or other drugs, or their adverse effect profile, as demonstrated here. Outside the clinic, such an analysis may be relevant to drug repurposing; occasionally, the unintended effects of medication under study for a given indication can form the basis for reapplication under a new indication. In a related application, drugs hitting a specific target can be queried against our clinical trial data to understand possible indications under investigation.
How are we joining these tables?
This query begins with a previously unexplored table as the starting point, structured_adverse_effects. As we are primarily interested in the specific conditions themselves, we first join it to adverse_effect_conditions on the 'id' of the adverse effect matching the 'adverse_effect_id' in the latter table. This table contains a 'condition_id,' which matches with the 'id' of the main conditions table. After this, we can join tables related to bio entities, bonds, and drugs, much as we did before.
Two important aspects of this query to note include the use of the SQL clause “LIKE,” which allows for rudimentary pattern matching. This allows us to capture the various 5-HT receptors with comparatively little query complexity. The other aspect to note is the filter on the 'relationship' of the adverse effect condition. Here, we filter to those rows where the value of this column is “effect.” This table also holds information related to conditions in other contexts of adverse effect information, such as characteristics of patient groups, which would not be relevant to this specific question.
Conclusion
For a wide variety of use cases, finding drug target information is a powerful way of identifying drugs of interest based on drug function. These drug actions may be desired or undesired; for the right question, both can help to narrow down a list of putative drugs of interest. The DrugBank database provides the power to interrogate these data and can exponentially strengthen your ability to search by making it easy to zoom in or out of drug groupings according to your desired topics such as:
- drug classification
- indications
- adverse effects
- drug approvals
- boxed warnings
- interactions
Drugs function at a molecular level by binding to distinct targets to elicit an effect; other interactions with enzymes, carriers, and transporters may be related to a drug's mechanism or may be important for its pharmacokinetic parameters. The DrugBank targets dataset, as laid out in the bonds table, provides an entry point into the DrugBank knowledgebase that reflects these interactions. Users can query this table to identify interactions directly for given drugs or drug categories. By joining additional tables in the knowledge base, it is possible to answer relevant questions for clinical practice, drug discovery and repurposing, and many other applications.